Affiliated member, Group Leader at Francis Crick Institute
Europe PMC | PubMed | CV & awards | Francis Crick Institute Profile | Goehring Lab
Twitter @GoehringLab
Research synopsis
One of the current grand challenges in biology is bridging the gap between detailed gene/interaction lists and phenotypic, systems-level behaviors of biological networks.
For patterning networks, molecular ensembles must sense and transduce spatial information such as position, direction and distance, which requires integrating information over length scales orders of magnitude larger than the molecules themselves. In most cases, we are only beginning to understand how networks accomplish such tasks, which will require probing and analyzing networks from the molecular to system scale.
My lab is taking on this challenge in the context of the conserved PAR polarity network. PAR proteins are essential transducers of spatial information between upstream symmetry-breaking cues and downstream pathways that control polarized processes such as cell migration, asymmetric cell division and tissue architecture. A core feature of this network is the segregation of two antagonistic groups of PAR proteins into opposing membrane domains. By analyzing this network across scales, we aim to identify how core design principles emerge from key molecular activities and behaviors, and in turn how these principles drive robust generation of polarity domains.
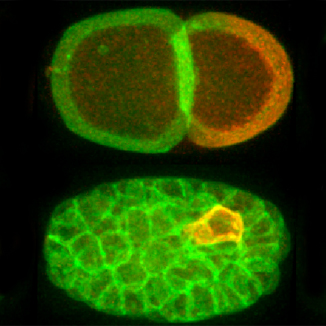
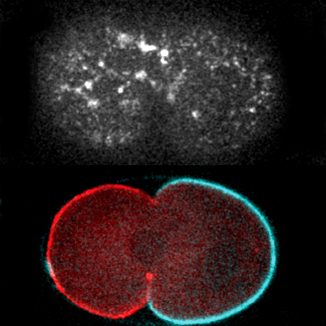
Selected publications
Rodriguez J*, Peglion FP*, et al (2017). aPKC cycles between functionally distinct PAR protein assemblies to drive cell polarity. Developmental Cell 42. *These authors contributed equally.
Trong PK, et al (2014). Parameter-space topology of models for cell polarity. New Journal of Physics, 16:065009.
Goehring NW, et al (2011). PAR proteins diffuse freely across the anterior-posterior boundary in polarized C. elegans embryos. J Cell Biol, 193:583-594.
Goehring NW, et al (2011). Polarization of PAR proteins by advective triggering of a pattern-forming system. Science, 334:1137-1141.
Goehring NW, et al (2010). FRAP analysis of membrane-associated proteins: lateral diffusion and membrane-cytoplasmic exchange. Biophys J, 99:2443-2452.
Funders
Medical Research Council
Wellcome Trust
Cancer Research UK
Horizon 2020 – European Commission
Research themes
Polarity and cell shape
Cytoskeleton and cell cortex
Signalling pathways
Technology
Light microscopy
People
Nisha Hirani (Senior Laboratory Research Scientist)
Florent Peglion (Postdoctoral Fellow)
Nelio Rodrigues (Postdoctoral Fellow)
Joana Pinto (Postdoctoral Fellow)
Lars Hubatsch (PhD Student)
Jake Reich (PhD Student)
Rukshala Illukkumbura (PhD Student)
Tom Bland (PhD Student)
 Close
Close


